Dr James Rosengarten reports highlights from the European Heart Rhythm Association (EHRA) Europace 2013 meeting held recently in Athens, Greece.
Details of two biomarker discovery programmes were presented by myself and other colleagues from Southampton.The early results were generated by our team, headed by Professor John Morgan, and build on several years of collaboration between cardiologists at the University Hospitals Southampton and scientists at the University of Southampton. The work hopes to advance sudden cardiac death risk stratification and ultimately move towards a more personal selection of interventions, such as implantable defibrillators.
Traditional risk stratification markers, such as left ventricular function or QRS width, fail to identify those at greatest risk, or merely demonstrate those with advanced heart failure who will go on to die regardless of what intervention is offered. This variation in response may, in part, be due to the genetic variability and response to, for example, ischaemic insult. Measuring this variability is challenging: gene expression does not accurately reflect functional protein expression. Proteomics is an emerging field in which this protein fingerprint can be sequenced directly.
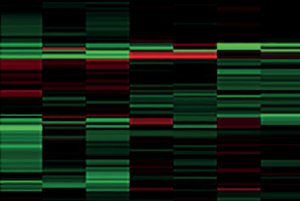
Our team have exploited these techniques, most frequently used in cancer sciences where solid tissue is readily available, to look for differences in protein expression of the whole serum proteome. We have taken serum samples from 243 patients with implantable cardiac devices, who were followed-up for 40 months for the occurrence of death, ventricular arrhythmia or survival without arrhythmia.1 Following pooling and sample preparation, mass spectrometry was used to demonstrate differential expression between groups (figure 1), generating over 90 proteins that were associated with arrhythmia and not survival.
Generating candidate biomarkers in this fashion means that whole panels of proteins can be rapidly selected for testing. Usually, serum biomarkers are chosen based upon only a limited understanding of cellular mechanisms, whereas this technique enables unbiased selection without preconceptions.
An emerging risk stratification tool in recent years has been the use of cardiac magnetic resonance imaging (MRI) to quantify myocardial scar. Although the association between myocardial scar and ventricular arrhythmogenesis has been well reported, cardiac MRI is resource and time limited, making it unsuitable as a screening tool.
Our Southampton group looked at whether the resting ECG could be a surrogate for myocardial scar but found traditional ECG parameters were poor at detecting scar accurately.2 Manual assessment of the ECG is time consuming and limited to those parameters that can be visually appraised. We know that the ECG signal contains much more information but extracting that data and processing large volumes requires a fresh approach. By working with biomedical signal engineers, the team sought to develop a novel algorithm that was capable of classifying scar. Standard 12-lead ECGs were recorded from over 150 patients undergoing cardiac MRI with scar assessment. Time, frequency and phase domain features were then ‘extracted’ from the digitally acquired signal and used in a ‘machine learning’ experiment to train the algorithm to classify ECGs with known scar burden (see figure 2). This classifier was then tested on the ECGs of over 80 patients; the algorithm correctly identified scar with 81% sensitivity and 73% specificity.
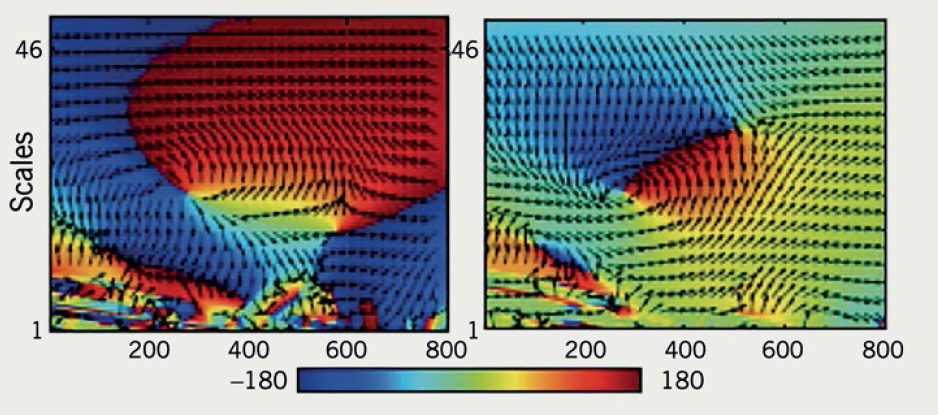
As more data are processed this type of approach can be endlessly refined. The classifier could be valuable in population screening before referral for more costly complex investigations, such as MRI scanning.
James Rosengarten
Wessex Electrophysiology Fellow and BJCA Deanery Representative
(james@rosengarten.co.uk)
References
- Rosengarten JA, Scott PA, Larkin SE et al. High resolution multidimensional proteomics detects candidate arrhythmia biomarkers. Presented at Europace 2013, Athens, Greece. Europace 2013;15(suppl 2):ii11. http://dx.doi.org/10.1093/europace/eut194
- Rosengarten JA, Dima SM, Panagiotou C et al. Novel non-invasive detection of arrhythmia substrate using supervised learning support vector machine. Presented at Europace 2013, Athens, Greece. Europace 2013;15(suppl 2):ii118. http://dx.doi.org/10.1136/heartjnl-2013-304019.72
