Infective endocarditis (IE) causes high rates of morbidity and mortality. Clinical management is problematic if there are uncertainties over the identity, viability or antibiotic susceptibility of the causative organism. Between 10% and 30% of IE blood cultures are negative, usually a result of prior antimicrobial therapy, but also occurring when causative micro-organisms are non-cultivable or fastidious. While evidence-based guidelines exist for treatment of IE caused by defined agents, clinicians are often faced with the dilemma of IE of unproven aetiology. Duration of empirical therapy is usually titrated against overall clinical response and non-specific laboratory markers of inflammation, but these may bear little relation to ongoing microbial activity in the heart valve. There is an increasing need for more specific, sensitive and rapid tests for the identification of causative organisms. Nucleic acid amplification technologies (NAATs) show promise for rapid detection of pathogen nucleic acid in blood or tissue. This review discusses the developments in this field, and the potential for the application of NAATs to improve aetiological identification in IE.
Introduction
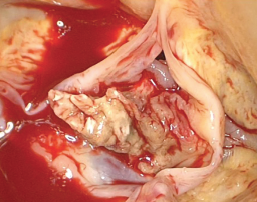
Untreated infective endocarditis (IE) is fatal; even with appropriate treatment, IE is associated with high rates of morbidity and mortality worldwide.1 The annual incidence of IE over the past two decades has remained relatively constant, ranging between 1.7 and 6.2 cases/100,000 population. Neither advances in healthcare nor revisions made to the current diagnostic criteria have substantially altered this.1-3 The current definition for IE now incorporates infections of prosthetic heart valves (both bioprosthetic and mechanical), implanted devices (such as pacemakers or ventricular assist devices) and cardiac endothelial surfaces.4 Figure 1 illustrates a large vegetation on the aortic valve from a patient with IE.
The variability in both the clinical manifestations and the course of IE reflect, in part, the heterogeneity of causative micro-organisms; this makes accurate diagnosis by clinical means alone problematic.5,6 Currently, the likelihood of IE is based on a score derived from a combination of clinical, microbiological and echocardiographic evidence.7-10 Laboratory diagnosis consists of culture of the infectious organism from the blood and/or heart valve material. However, the sensitivity of this scoring system is significantly compromised when IE is caused by fastidious or non-cultivable organisms or when patients have received previous antibiotic therapy such that positive cultures cannot be obtained. Novel diagnostic tests, such as nucleic acid amplification technologies (NAATs), that can identify the presence of infective organisms irrespective of the culture constraints will not only improve sensitivity but may reduce empiric treatment by permitting targeted antibiotic therapy. To be clinically useful any such technique must be able to identify the causative micro-organism from blood or tissue rapidly, have appropriately high positive- and negative-predictive values, and demonstrate good reproducibility between laboratories.
This review discusses the developments and application of NAATs for rapid and accurate detection of the infectious aetiology of IE.
Epidemiology and microbiology of IE
IE affects both native and prosthetic (bioprosthetic and mechanical) valves, and may develop through community- or healthcare-associated acquisition (CA-IE and HCA-IE, respectively). Worldwide, CA-IE of the native valve is by far the most common form of IE with rheumatic valve disease remaining the major risk factor;4 in resource-rich countries, however, profound changes in the epidemiology and aetiology of IE have been seen in recent years. There are many reasons for such shifts, but the principles for IE risk remain essentially unaltered: increased opportunity for microbial entry to the blood circulation, the presence of abnormal endocardial surfaces or flow patterns, and diminished host immune capacity.3,4,11 In developed countries with expanding elderly populations, degenerative valve lesions and congenital defects have far outstripped chronic rheumatic valve disease as major underlying risk factors, being present in up to 50% of IE patients over the age of 60 years.5
Although native valve endocarditis remains mostly CA-IE, the incidence of HCA-IE is steadily increasing12 as medical interventions allow greater opportunities for microbial access to the bloodstream via prolonged or repeated intravascular access or cannulation. Cardiothoracic surgical advances in developed healthcare settings have provided the emergence of new risk groups, including patients with prosthetic valves, intravascular devices or endovascular repairs. The flora associated with HCA-IE, predominantly skin-dwelling staphylococcal species, is fundamentally different to that of CA-IE. Such differences therefore demand different treatment.13,14 IE associated with intravenous drug use (IVDU) is in many ways unique – left- and right-sided valves are affected in approximately equal proportions,14 and the variety of microbial flora reflects the different opportunities for contamination at different stages of the process.15,16 Staphylococcus aureus is the predominant organism of IVDU-related IE as it colonises skin flexures; IE caused by Pseudomonas species and other motile Gram-negative bacteria have been attributed to the use of contaminated water to clean needles,17 and dissolving heroin in lemon juice predisposes to candidaemia and Candida IE.18
Prosthetic valve endocarditis occurs at a rate of 3–6/1,000 patient-years, accounting for an estimated 1–5% of all IE cases in resource-rich settings. Early prosthetic valve endocarditis occurs within 60 days of valve surgery11 and is typically HCA-IE, with Staph. aureus, Staph. epidermidis and other coagulase-negative staphylococci the most common pathogens. By contrast, the microbiology of late prosthetic valve endocarditis resembles that of native valve CA-IE.19 The typical microbiology of IE and its patterns of acquisition are summarised in table 1 (available in the PDF download).
Current diagnostic methods used in IE
The recognised epidemiological characteristics, described above, are only associations, and, while they may guide empiric therapy, they are not sufficiently reliable to allow antimicrobial prescribing in individual cases with certainty. For this, microbiological identification of the causative organism is needed; this will also inform the decisions on duration of therapy and even whether surgical intervention should be anticipated.20
However, direct microbiological culture from blood is not without pitfalls. The protean manifestations of IE that in part reflect the variety of aetiological agents, mean that patients frequently present to primary care with non-specific symptoms. Bacteraemia associated with IE is usually continuous but low grade, averaging at 1–10 organisms per ml of blood;21,22 blind trials of antibiotic therapy may, therefore, suppress microbial activity at the valve sufficiently to reduce the sensitivity of blood cultures taken at the time of hospital presentation.2,20-23 Indeed, up to 30% of all IE cases have negative blood cultures.24 A minor proportion of these cases are due to the presence of micro-organisms that are either non-cultivable, fastidious in their nutritional requirements in culture, or are extremely slow-growing on conventional media. The modified Duke criteria try to compensate for these issues, but the scoring system remains flawed (table 2 available in the PDF download).
Rationale for use of NAATs in IE
The advent of molecular technology, the “diagnostic tool for the new millennium”,25 has turned opportunities for rapid organism identification and even detection of certain drug susceptibility patterns from concept into a working reality. The rapid exponential generation of billions of copies of target DNA template from a single original sequence accounts for the potential for NAATs to be sensitive, specific and timely. One of the greatest assets of molecular tests is their potential to be adapted according to purpose. Amplification tests may be designed to be so specific as to detect only a single species, strain, or even resistance-inducing mutation; alternatively, the use of commonly shared genetic sequences as amplification targets allows detection of much broader categories of organisms. For example, the 16S rDNA gene codes for the RNA component of the 30S sub-unit of the prokaryotic ribosome. As it has both highly conserved and variable regions, pan-bacterial primers can be developed to target the conserved regions that immediately adjoin variable regions. Consequently, a single set of primers can be used to amplify the DNA from an enormous range of different bacteria. Subsequent sequencing of this amplified DNA can identify the variable region and thereby the bacterium.26-28 As the 16S rDNA gene sequence is universal throughout all phyla of bacteria it is, therefore, ideally suited for the diagnostics of IE, where there is such immense diversity of possible causative organisms including those that are fastidious or non-cultivable, or even no longer viable.3,29 The majority of NAATs applied to IE have used polymerase chain reaction (PCR) to amplify and subsequently sequence the 16S rDNA gene. To date, this technique has been used most successfully on excised valve tissue, and at present cannot replace microbiological culture of blood. Table 3 ( available in the PDF download) outlines the potential advantages and disadvantages of using molecular techniques to diagnose IE, with specific regard to targeting the 16S rDNA gene.
In fungal endocarditis, the 18S rDNA gene – the equivalent of the bacterial 16S rDNA gene – is more problematic as a diagnostic target since 18S rDNA gene sequences are highly conserved and demonstrate insufficient variability to differentiate between many fungal species. Instead, alternative targets such as short non-coding ribosomal internal transcribed spacer (ITS) regions are increasingly being used. These regions are located between conserved genes encoding for 18S, 5.8S and 28S rDNA, and are highly variable in both length and sequence, and, thus, more efficient for discriminating species than 18S sequences.30
Real-time PCR
More recently, 16S real-time or quantitative PCR (qPCR) has been applied to bacterial IE.2,31 There are a number of advantages of qPCR over conventional PCR: not only is it a more sensitive technique, but it is also more rapid as it eliminates the need for post amplification steps such as gel electrophoresis of PCR products.11 Importantly, qPCR can also measure the amount of inhibition from clinical samples and, hence, evaluate the effectiveness of the nucleic acid extraction method. For this, internal extraction and amplification controls are added to the sample before each of these steps.32
Previous studies have recommended that molecular-based techniques are included as a major criterion in the Duke criteria.22,31,33 Indeed, several studies have now demonstrated PCR positivity on valves in patients classified as possible or definitive IE, even when blood culture was negative.3 Results from these studies are undoubtedly promising, with the sensitivity of PCR from valve material ranging from 41.2% to 96%, compared with direct culture rates of 7.8–24.3%. Information of all published trials using 16S rDNA PCR in IE is available in a supplementary appendix (available online at www.bjcardio.co.uk).
Conclusions
It is important to remember that whatever the method of organism identification in IE, whether culture isolation or molecular nucleic acid detection, the result must still be open to careful interpretation. With an ever-increasing list of organisms that have been associated with both native and prosthetic valve IE, this assessment becomes more complex. Attribution of causation must always be weighed against the possibility of contamination. Appropriate measures to prevent contamination are as critically important in microbiological culture as in the molecular laboratory.
Molecular tests undoubtedly advance the diagnosis of IE; however, a much greater understanding of the variables that influence the sensitivities and specificities of the molecular methods needs to be defined. Recent calls to introduce basic standards in molecular diagnostic test protocols,34 in conjunction with the use of common targets, would allow for a much more accurate comparison between studies. With advances in molecular technology, NAATs now provide far more sensitive and rapid methods to detect the micro-organisms that cause IE. Comparative results from recent studies using NAATs indicate the clear superiority of valve material PCR over conventional valve culture. PCR can provide a positive identification where one or more of the definitive Duke criteria have been inconclusive. Development of consensus guidelines is needed to overcome the difficulties that the lack of standardisation of targets and protocols present to enabling valid comparisons between studies. With recent advances in nucleic acid quantification, NAATs technology may provide a tool to help answer some of the outstanding challenges that remain for IE diagnostics: accurate treatment response monitoring and reliable outcome prediction.
Acknowledgement
SM-J and AZ receive support from the UK Comprehensive Biomedical Research Centre and the National Institue of Medical Research.
Conflict of interest
None declared.
Editors’ note
A supplementary appendix containing information on all published trials using 16S rDNA PCR in IE is available online at www.bjcardio.co.uk
Key messages
- Infective endocarditis (IE) causes high rates of morbidity and mortality
- Clinical management is problematic if there are uncertainties over the identity, viability or antibiotic susceptibility of the causative organism
- Conventional diagnostic microbiological techniques fail when patients have received prior antibiotic therapy, or when the causative organism is fastidious or non-cultivable
- Nucleic acid amplification techniques (NAATs) now represent a much more sensitive and rapid method for detection of the micro-organisms that cause IE compared with culture alone
References
1. Prendergast B. The changing face of infective endocarditis. Heart 2006;92:879–85.
2. Munoz P, Bouza E, Marin M et al. Heart valves should not be routinely cultured. J Clin Microbiol 2008;46:2897–901.
3. Tak T. Molecular diagnosis of infective endocarditis: a helpful addition to the Duke criteria. Clin Med Res 2004;2:206–08.
4. Bashore T, Cabell C, Fowler V. Update on infective endocarditis. Curr Probl Cardiol 2006;31:274–352.
5. Moreillon P. Infective endocarditis. Lancet 2004;363:139–49.
6. Millar B, Moore J. Antibiotic prophylaxis, body piercing and infective endocarditis. J Antimicrob Chemother 2004;53:123–6.
7. Li J, Sexton D, Mick N et al. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin Infect Dis 2000;30:633–8.
8. Lamas C, Eykyn S. Suggested modifications to the Duke criteria for the clinical diagnosis of native valve and prosthetic valve endocarditis: analysis of 118 pathologically proven cases. Clin Infect Dis 1997;25:713–19.
9. Fournier P-E, Casalta J, Habib G, Messana T, Raoult D. Modification of the diagnostic criteria proposed by the Duke Endocarditis Service to permit improved diagnosis of Q fever endocarditis. Am J Med 1996;100:629–33.
10. Bayer AS, Bolger AF, Taubert KA et al. Diagnosis and management of infective endocarditis and its complications. Circulation 1998;98:2936–48.
11. Syed F, Millar B, Prendergast B. Molecular technology in context: a current review of diagnosis and management of infective endocarditis. Prog Cardiovasc Dis 2007;50:181–97.
12. Eykyn S. Endocarditis: basics. Heart 2001;86:476–80.
13. Cosgrove S, Karchmer A. Endocarditis. Medicine 2005;33:66–72.
14. Fowler V, Miro J, Hoen B. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA 2005;293:3012–21.
15. Brook I. Infective endocarditis caused by anaerobic bacteria. Arch Cardiovasc Dis 2008;101:665–76.
16. Baddour L, Wilson W, Bayer A et al. Infective endocarditis: diagnosis, antimicrobial therapy, and management of complications: a statement for healthcare professionals from the Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, and the Councils on Clinical Cardiology, Stroke, and Cardiovascular Surgery and Anesthesia, American Heart Association: Endorsed by the Infectious Diseases Society of America. Circulation 2005;111:e394–e434.
17. Oh S, Havlen P, Hussain N. A case of polymicrobial endocarditis due to anaerobic organisms in an injection drug user. J Gen Intern Med 2005;20:C1–C2.
18. Bisbe J, Miro J, Latorre X et al. Disseminated candidiasis in addicts who use brown heroin: report of 83 cases and review. Clin Infect Dis 1992;15:910–23.
19. Loupa C, Mentzikof D. Current opinions in infective endocarditis. Hellenic J Cardiol 2002;43:53–62.
20. Voldstedlund M, Pedersen L, Baandrup U, Klaaborg K, Fuursted K. Broad-range PCR and sequencing in routine diagnosis of infective endocarditis. APMIS 2008;116:190–8.
21. Watkin R, Lang S, Lambert P, Littler W, Elliott T. The microbial diagnosis of infective endocarditis. J Infect 2003;47:1–11.
22. Millar B, Moore J, Mallon P et al. Molecular diagnosis of infective endocarditis – a new Duke’s criterion. Scand J Infect Dis 2001;33:673–80.
23. Beynon R, Bahl V, Prendergast B. Infective endocarditis. BMJ 2006;333:334–9.
24. Naber C, Erbel R. Infective endocarditis with negative blood cultures. Int J Antimicrob Agents 2007;30S:S32–S36.
25. Yang S, Rothman R. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis 2004;4:337–48.
26. Barken K, Haagensen J, Tolker-Nielsen T. Advances in nucleic acid-based diagnostics of bacterial infections. Clin Chim Acta 2007;384:1–11.
27. Rice PA, Madico GE. Polymerase chain reaction to diagnose infective endocarditis. Will it replace blood cultures? Circulation 2005;111:1352–4.
28. Petti C. Detection and identification of microorganisms by gene amplification and sequencing. Clin Infect Dis 2007;44:1108–14.
29. Clarridge III J. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev 2004;17:840–62.
30. Deng W, Xi D, Mao H, Wanapat M. The use of molecular techniques based on ribosomal RNA and DNA for rumen microbial ecosystem studies: a review. Mol Biol Rep 2008;35:265–74.
31. Marin M, Munoz P, Sanchez M et al. Molecular diagnosis of infective endocarditis by real-time broad-range polymerase chain reaction (PCR) and sequencing directly from heart valve tissue. Medicine 2007;86:195–202.
32. Mackay I. Real-time PCR in the microbiology laboratory. Clin Microbiol Infect 2004;10:190–212.
33. Bosshard P, Kronenberg A, Zbinden R, Ruef C, Bottger E, Altwegg M. Etiologic diagnosis of infective endocarditis by broad-range polymerase chain reaction: a 3-year experience. Clin Infect Dis 2003;37:167–72.
34. Bustin S, Benes V, Garson J et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009;55:611–22.
35. Mylonakis E, Calderwood S. Infective endocarditis in adults. N Engl J Med 2001;345:1318–30.
36. Brouqui P, Raoult D. Endocarditis due to rare and fastidious bacteria. Clin Microbiol Rev 2001;14:177–207.
37. Mandell G, Bennett J, Dolin R. Cardiovascular infections. In: Principles and practice of infectious diseases. Philadelphia: Elsevier/Churchill Livingstone, 2005.
38. Cuculi F, Toggweiler S, Auer M, Auf der Maur C, Zuber M, Erne P. Serum procalcitonin has the potential to identify Staphylococcus aureus endocarditis. Eur J Clin Microbiol Infect Dis 2008;27:1145–9.
39. Dreier J, Vollmer T, Freytag C, Baumer D, Korfer R, Kleesiek K. Culture-negative infectious endocarditis caused by Bartonella spp.: 2 case reports and a review of the literature. Diagn Microbiol Infect Dis 2008;61:476–83.
40. Lang S, Watkin RW, Lambert PA, Bonser RS, Littler WA, Elliott TSJ. Evaluation of PCR in the molecular diagnosis of endocarditis. J Infect 2004;48:269–75.
41. Mueller C, Huber P, Laifer G, Mueller B, Perruchoud A. Procalcitonin and the early diagnosis of infective endocarditis. Circulation 2004;109:1707–10.
42. Christ-Crain M, Muller B. Procalcitonin in bacterial infections – hype, hope, more or less? Swiss Med Wkly 2005;135:451–60.
43. Hodgetts A, Levin M, Kroll J, Langford P. Biomarker discovery in infectious diseases using SELDI. Future Microbiol 2007;2:35–49.
44. Sontakke S, Cadenas M, Maggi R, Diniz P, Breitschwerdt E. Use of broad-range 16S rDNA PCR in clinical microbiology. J Microbiol Methods 2009;76:217–25.
45. Millar B, Moore J. Current trends in the molecular diagnosis of infective endocarditis. Eur J Clin Microbiol Infect Dis 2004;23:353–65.
46. Johnson J. Development of polymerase chain reaction-based assays for bacterial gene detection. J Microbiol Methods 2000;41:201–09.
47. Silkie S, Tolcher M, Nelson K. Reagent decontamination to eliminate false-positives in Escherichia coli qPCR. J Microbiol Methods 2008;72:275–82.
48. Podglajen I, Bellery F, Poyart C et al. Comparative molecular and microbiologic diagnosis of bacterial endocarditis. Emerg Infect Dis 2003;9:1543–7.
49. Haanpera M, Jalava J, Huovinen P, Meurman O, Rantakokko-Jalava K. Identification of alpha-haemolytic streptococci by pyrosequencing the 16S rRNA gene and by use of VITEK 2. J Clin Microbiol 2007;45:762–70.
50. Ghebremedhin B, Layer F, Konig W, Konig B. Genetic classification and distinguishing of Staphylococcus species based on different partial gap, 16S rRNA, hsp60, rpoB, sodA and tuf gene sequences. J Clin Microbiol 2008;46:1019–25.
51. Faibis F, Mihaila L, Perna S et al. Streptococcus sinensis: an emerging agent of infective endocarditis. J Med Microbiol 2008;57:528–31.
52. Fihman V, Raskine L, Barrou Z et al. Lactococcus garvieae endocarditis: identification by 16S rRNA and sodA sequence analysis. J Infect 2006;52:e3–e6.
53. Huggett J, Novak T, Garson J et al. Differential susceptibility of PCR reactions to inhibitors: an important and unrecognised phenomenon. BMC Research Notes 2008;1(1):70.
